Light has always been the best physical tool to observe structures and molecules in cell biology. But recently, cell biology has gone through a technical revolution by using light and photosensitive proteins to control molecular mechanisms in cells and to measure physico-chemical parameters such as membrane tension. Here, the authors report on the recent development of a promising chemical tool and a powerful informatics tool for measuring physical cell parameters with light.
Optogenetics tools allow for protein depletion in the cytoplasm, induction or disruption of protein interaction on demand. Moreover, chemical biology tools to measure forces occurring at the molecular scale have also bloomed, in particular in studies of the cytoskeleton. Here, we describe the recent development of a promising chemical tool and a powerful informatics tool for measuring physical cell parameters with light.
Within the NCCR Chemical Biology, conceptually new approaches to fluorescent membrane probes have been developed by the Matile’s group (UNIGE). Adai Colom, in the Roux group (UNIGE), used such a fluorescent probe named FliptR (Fluorescent LIPid Tension Reporter) (1) to measure plasma membrane tension within living cells (Colom et al., under revision). FliptR is a planarizable push-pull probe that reports applied forces through changes of its fluorescence parameters, i.e. absorbance and emission peaks, intensity and lifetime.
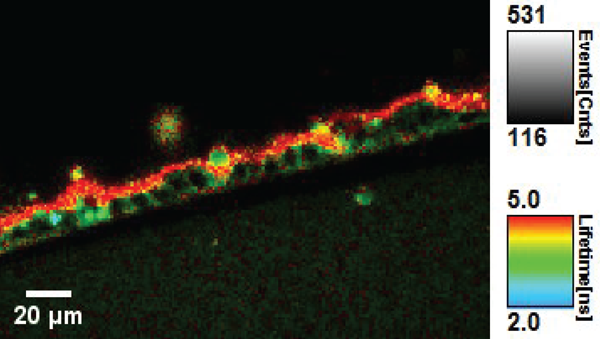
The rationale behind this approach is that in the confined environment of a lipid bilayer, FliptR is subjected to compressive forces that promote mechanical ground-state planarization, allowing for intramolecular charge transfer and increase of the fluorescence lifetime. Thus, FliptR is theoretically sensitive to different lipid phases and compositions, as well as to membrane tension, as all affect the compressive forces exerted within the bilayer.
We used fluorescence lifetime imaging microscopy (FLIM) to measure the change of fluorescence lifetime of FliptR on lipid phases and membrane tension, as a reflection of the lipid packing. The fluorescence lifetime of the probe is independent of the fluorescence intensity, which changes by a factor of 10 in the planarized state versus the twisted one. The fluorescence lifetime is thus also independent of the optical set-up used to measure it. We showed that the lifetime of FliptR depends linearly on membrane tension within cells, allowing for an easy quantification of the membrane tension.
Hence, the FliptR probe provides an interesting and polyvalent tool for measuring membrane tension in the plasma and intracellular membrane compartments. Mechanical parameters associated with cells can be dramatically affected in disease conditions, e.g. cancer cells are known to be stiffer, so tools such as our FliptR probe can be potentially useful and allow for example to visualize and sort out cancer cells in blood samples or directly in the body of patients.

Another application related to the use of light is the development of a software for the acquisition of 3D image datasets – a new type of information about cells – which is now a common practice in biology.
Our particular interests in the lab include the study of epithelium mechanics, where the shape of cells has to be well described in 3D terms with data sets originated from images produced by either electron microscopy or fluorescent confocal microscopy, both approaches containing useful information on cell shape. Acquiring such data is a relatively easy part but extracting real 3D cell shape information from these data can be challenging. A way to achieve this requires state-of-the-art bioimaging analysis, recently boosted by the notable irruption of capabilities offered by the developments in machine learning in general and convolutional neural network in particular. To fully harness the power of these methods in a biochemistry lab can be difficult with the limited expertise in bioinformatics, so there is a need for easy-to-use software tools. The famous and constantly evolving ImageJ/FIJI software is such an example but there is still room for development.

Within this context, a new 3D segmentation tool was created in the Biochemistry Department (UNIGE) and proved to be both user-friendly and meet the particular needs of biologists. The method called LimeSeg (Lipid Membrane Segmentation) is loosely based on a simulation of lipid membrane whereby the surface is implicitly defined by sets of shortly-coupled particles, resembling lipid particles. As part of the family of active contour methods, the user defines one or several initial shapes, generally spheres, that are attracted by local image maxima. The multiple seeds evolve until they reach the threshold of objects, like cells in an epithelium. This approach proved very useful in various contexts such as fluorescence microscopy, EM and even MRI, while offering a very good compromise in terms of outcomes and user friendliness. The Roux lab is now working to include LimeSeg in the very popular ImageJ/Fiji software (Machado, 2018) so that it becomes freely available to all the community.
References
- S. Soleimanpour et al., Headgroup engineering in mechanosensitive membrane probes, Chem. Commun. (Camb.), 2016.
- M. Dal Molin et al., Fluorescent flippers for mechanosensitive membrane probes. J. Am. Chem. Soc., 2015.
- F. Neuhaus et al., Correlation of surface pressure and hue of planarizable push–pull chromophores at the air/water interface, Beilstein Journal of Organic Chemistry, 2017.
Article’s authors: Caterina Tomba, Adai Colom Diego, Nicolas Chiaruttini and Aurélien Roux, Biochemistry Department, University of Geneva

Leave a comment
The editors reserve the right not to publish comments or to abridge them.