Single-molecule imaging approaches allow today’s researchers to follow the movement, interactions and conformational dynamics of single molecules in real-time, thus to provide novel insights into complex biochemical systems that have remained masked in traditional averaging data approaches. In this article, Prof. Fierz explains how the combination of such fluorescence imaging methods provides an unprecedented view of chromatin dynamics.
Cellular processes involve the collective action of large ensembles of macromolecules, resulting in emergent properties that cannot be gleaned from the study of individual molecules. The ability to observe individual molecules within such complex ensembles however provides novel means to study dynamic biochemical processes.
In the Fierz lab, we study the function of chromatin, the nucleoprotein complex organizing the eukaryotic genome. The molecular components of chromatin include the histone proteins and DNA segments making up nucleosomes, as well as the associated architectural and regulatory proteins. Together, they form a complex system controlling gene expression, cell inheritance and differentiation. Moreover, chromatin is highly dynamic and structurally heterogeneous, rendering detailed molecular studies difficult. Thus, we develop simplified model systems combining protein chemistry (to assemble defined chromatin segments) with single-molecule fluorescence measurements. Together these methods allow us to observe chromatin structure, dynamics and biochemistry in mechanistic detail.
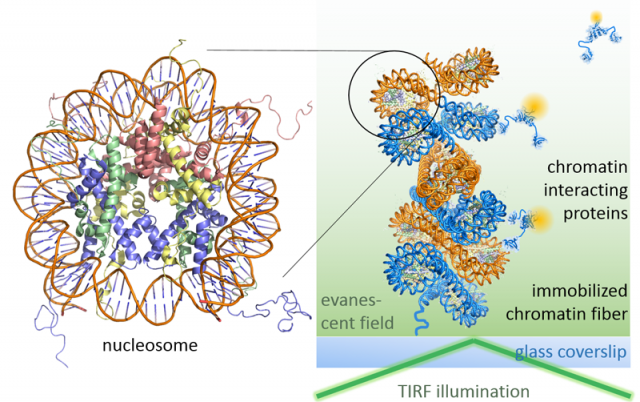
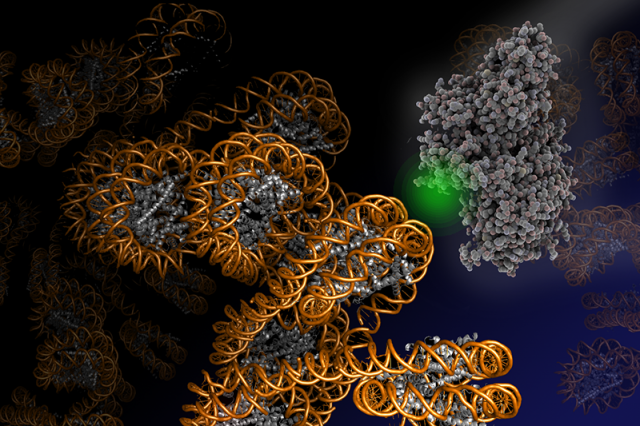
Leveraging fluorescence methods, we have recently established two complementary approaches to dissect chromatin biochemistry on the single-molecule level. A first approach, colocalization microscopy (Figure 1), allows us to directly watch regulatory proteins while they interact with and act upon chromatin segments. For such visual biochemistry experiments, we immobilize synthetically produced and fluorescently labeled chromatin fibers in a microfluidic flow cell. Subsequently, we inject the regulatory protein of interest (labeled with a fluorescent dye of different color) into the flow cell. Imaged under total internal reflection fluorescence (TIRF) conditions, transiently chromatin-bound protein appear as diffraction-limited spots that perfectly overlap with the fluorescence emission arising from immobilized chromatin fibers. TIRF microscopy only reveals molecules that remain close to the surface of the flow cell over the camera integration time (which is in the millisecond regime). Therefore, unbound proteins, which rapidly diffuse through the flow cell far from the illuminated surface, remain invisible. We then record movies which directly show the proteins interacting with chromatin, dependent on their functional state, chromatin structure and epigenetic modifications present. This approach thus allowed us to discover molecular principles governing the establishment of transcriptionally silent chromatin. Moreover, we could determine how PRC2, a key epigenetic regulatory factor that controls the expression of developmental genes, is recruited to its target sites and acts upon chromatin (Figure 2). In ongoing studies, we now apply this versatile method to decipher how transcription factors invade compact chromatin fibers, and how chromatin is remodeled during DNA damage repair.
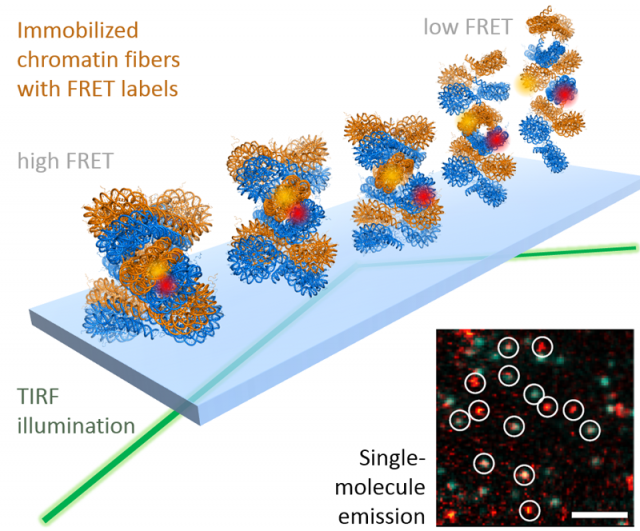
While colocalization microscopy yields kinetic and mechanistic information on the interaction of proteins with the chromatin, it yields no information on dynamic structural changes in chromatin associated with biological processes. Such knowledge is however of great importance for a deeper understanding of how genes are activated or how DNA damage is repaired. To gain real-time insight into chromatin structural transition, we have thus developed a suite of single-molecule Förster resonance energy transfer (FRET) methods. FRET provides information about the molecular distance (potentially with Angstrom precision) between two fluorescent labels placed within a biomolecule, for example a chromatin fiber. Using such a single-molecule FRET approach, we could recently directly measure how chromatin fibers move (Figure 3). This dynamic structural biology study reveals that, despite their large size in molecular terms, chromatin fibers exhibit multiscale motions in the micro- and millisecond time regime. Importantly, these motions persist in compact, transcriptionally silent chromatin states that before have been believed to be inert and static. We believe that such intrinsic motions allow chromatin fibers to quickly react to biological stimuli and allow DNA access to regulatory proteins. We are now systematically exploring chromatin dynamics in different biological processes with a focus on gene expression and DNA repair.
Together, our combined fluorescence imaging methods provide an unprecedented view of chromatin dynamics. We believe that the combined power of visual biochemistry and dynamic structural biology approaches will reveal molecular detail of chromatin regulatory and epigenetic processes. Due to the importance of these regulatory pathways for cell function and disease, molecular and mechanistic information is highly valuable for both a basic understanding of eukaryotic life and for the development of novel therapeutics. Single-molecule fluorescence methods, such as developed by our group, are thus a key part of the investigative toolkit to provide such information, together with chemical biology, biochemistry, cell biology and genetics approaches.
Readings
- An overview on chromatin dynamics: Cuvier, O. & Fierz, B., Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells, Nat. Rev. Genet., 2017.
- Single-molecule FRET: Kilic, S., Felekyan, S., Doroshenko, O., Boichenko, I., Dimura, M., Vardanyan, H., Bryan, L.C., Arya, G., Seidel, C.A.M. & Fierz, B., Single-molecule FRET reveals multiscale chromatin dynamics modulated by HP1alpha, Nat. Commun., 2018.
- Colocalization microscopy: Choi, J., Bachmann, A.L., Tauscher, K., Benda, C., Fierz, B. & Muller, J., DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation, Nat. Struct. Mol. Biol., 2017.
- Colocalization microscopy: Kilic, S., Bachmann, A.L., Bryan, L.C. & Fierz, B., Multivalency governs HP1alpha association dynamics with the silent chromatin state, Nat. Commun., 2015.

Leave a comment
The editors reserve the right not to publish comments or to abridge them.