The next revolution in microscopy is here. High-throughput imaging is the most efficient and rapid technique for the acquisition and analysis of a large number of images to achieve unbiased discovery of complex and precise cellular mechanisms.
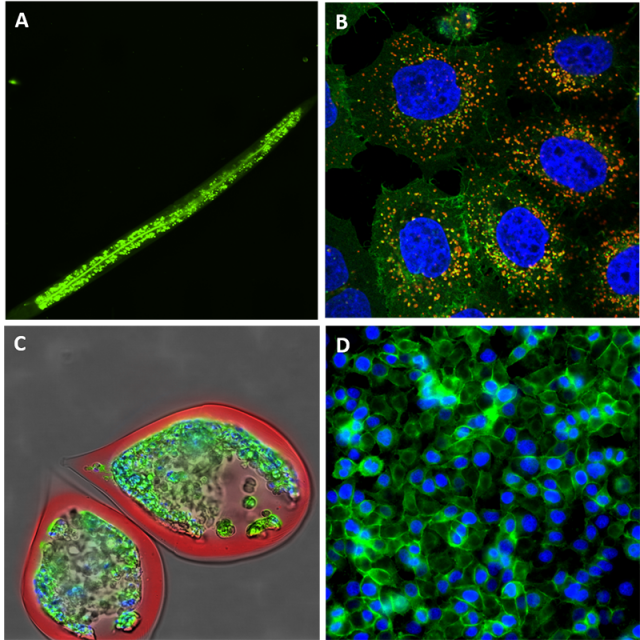
The invention, in the early 90s, of the automated fluorescence microscope opened totally new ways of screening biological processes. This new tool allows acquisition of many images without human intervention, images that can be used for large-scale screens. Combined with the increasing development of fluorescent cellular markers, the technology became rapidly the most efficient technique to run systems biology projects on complex and precise cellular processes. Today, thanks to the variety of antibodies or fluorescent chemicals (eg. Lysotracker®, mitotracker®), we can virtually image any cellular organelle or component [1] and quantify precisely any phenotype occurring in a chemical or genetic screening campaign. It is now possible to design much more precise assays, to allow for the identification and selection of small molecules with a specific activity on the biological process under study. Phenotypic screening became the method of choice for the discovery of target-specific drug candidates [2, 3].
In the academic environment, high-throughput fluorescence microscopy becomes rapidly a very powerful technique for deep systems biology analysis of any cellular pathways [4, 5]. Even more, it seems clear that automated fluorescence microscopy, thanks to the increasing quality of the hardware, will soon replace regular microscopy. The main advantages being the acquisition speed, the increasing number of samples imaged and analyzed, and the fully unbiased way of acquiring the images. All these features allow the labs to increase the number of experimental conditions and especially increase the confidence one can have in the detection and quantification of any, even very mild, phenotype [6].
Fluorescence microscopy involves staining of the sample. The high-throughput imaging is incompatible with manual work. Therefore, we use at ACCESS Geneva, dedicated robotic tools to stain and wash samples. In addition to time saving, this automated procedure allows for a very fast and consistent staining and, thanks to the miniaturization, a notable cost saving on the reagent can be achieved. These robotic assisted methods ensure also a much easier repeatability of the experiment along time.
Originally, automated fluorescence microscopy allowed only for 2D-fixed imaging, but today the technology has drastically evolved to 3D live imaging so that even full imaging of small organisms (see figure) is very commonly performed [7]. Fluorescence imaging allow the tracking of cells, organelles, protein complexes and even RNA molecules [8]. We can multiplex the colours within the limit of the overlapping wavelengths and use part of the channels for localization purposes and part for the tracking of the biological process.
In addition to drug discovery and fundamental cell biology, high-throughput fluorescence microscopy has ushered in the growing area of personalized medicine [9–10–11]. Indeed, the technology could be very useful to help finding the proper cure for a specific patient as it is possible to screen a set of drugs on a patient derived cell culture (mainly tumour-derived cells, cultured in 3D spheroid, to be as close as possible to the natural tumoral environment). This way, one can get an immediate answer about the potency of any given treatment for a specific patient. This technology is still tested over several institutes and hospitals around the world, but it seems very clear that it will play an important role in the tailoring of treatment to the molecular or genetic mapping of individual patients thus in personalized/precision medicine which holds the future of medicine.
References
- Wallabregue, A., et al., Selective Imaging of Late Endosomes with a pH-Sensitive Diazaoxatriangulene Fluorescent Probe, J. Am. Chem. Soc., 2016.
- Zanella, F., J.B. Lorens, and W. Link, High content screening: seeing is believing, Trends Biotechnol., 2010.
- Moffat, J.G., J. Rudolph, and D. Bailey, Phenotypic screening in cancer drug discovery – past, present and future, Nat. Rev. Drug Discov., 2014.
- Moreau, D. and J. Gruenberg, Automated Microscopy and High Content Screens (Phenotypic Screens) in Academia Labs, Chimia Int. J. for Chemistry, 2016.
- Chia, J., et al., RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells, Mol. Syst. Biol., 2012.
- Mattiazzi Usaj, M., et al., High-Content Screening for Quantitative Cell Biology, Trends Cell. Biol., 2016.
- Sirenko, O., et al., Phenotypic Characterization of Toxic Compound Effects on Liver Spheroids Derived from iPSC Using Confocal Imaging and Three-Dimensional Image Analysis, Assay Drug Dev Technol, 2016.
- Querido, E., L. Dekakra-Bellili, and P. Chartrand, RNA fluorescence in situ hybridization for high-content screening, Methods, 2017.
- Fong, E.L.S., et al., 3D Culture as a Clinically Relevant Model for Personalized Medicine, SLAS Technol, 2017.
- Vilja, P., et al., The High Throughput Biomedicine Unit at the Institute for Molecular Medicine Finland: High Throughput Screening Meets Precision Medicine, Combinatorial Chemistry & High Throughput Screening, 2014.
- Yu, K.K., et al., High content screening of patient-derived cell lines highlights the potential of non-standard chemotherapeutic agents for the treatment of glioblastoma, PLoS One, 2018.

Leave a comment
The editors reserve the right not to publish comments or to abridge them.